婴儿胆汁淤积的肠道菌群组成及功能变化
DOI: 10.3969/j.issn.1001-5256.2021.01.025
Composition and functional change of intestinal microbiota in infantile cholestasis
-
摘要:
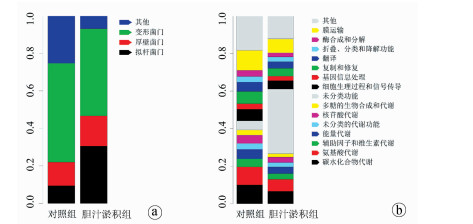
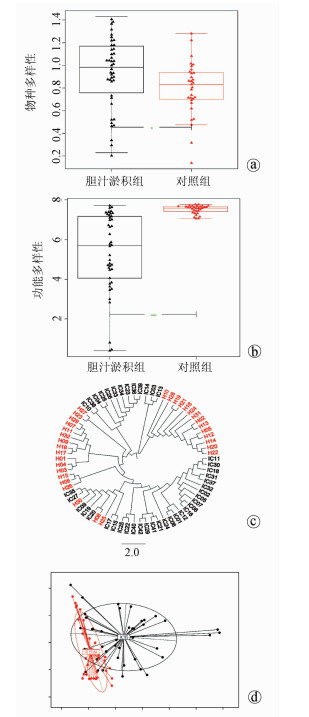
目的 研究胆汁淤积婴儿的肠道菌群组成及功能变化。 方法 纳入2014年9月—2016年2月深圳市儿童医院收治的42例胆汁淤积婴儿(胆汁淤积组)及同期32例正常对照婴儿(对照组),收集以上婴儿的大便标本,采用鸟枪法测序了解入选婴儿肠道菌群特征,应用京都基因与基因组百科全书、eggNOG数据库探索菌群功能改变。计量资料2组间比较采用Wilcoxon秩和检验; 计数资料2组间比较采用χ2检验。得到的菌种和基因功能通路等数据用Shannon系数、Wilcoxon秩和检验和DESeq2进行计算分析,利用Cytoscape将2组的丰富菌种和相关联系直观表现出来。 结果 胆汁淤积组病因包括胆道闭锁11例(26.2%),Citrin蛋白缺乏症4例(9.5%),胆管发育不良4例(9.5%),原因未明23例(54.8%)。2组肠道菌群组成在门水平有显著差异(Z=0.000 046,P<0.05),其中厚壁菌门和拟杆菌门在胆汁淤积组中显著增加; 在种水平上也有差异(Z=0.000 014,P<0.05),胆汁淤积组中有益菌如双歧杆菌明显减少,而潜在致病菌如韦永氏球菌和农研丝杆菌明显增多; 在功能水平上也有差异(Z=0.019 8,P<0.05),胆汁淤积组肠道菌群的功能明显减弱,如氨基酸代谢、糖代谢等。 结论 胆汁淤积婴儿肠道菌群的组成及功能发生改变,胆汁淤积婴儿肠道菌群中有益菌明显减少,潜在致病菌增多,补充双歧杆菌等有益菌可能减少潜在致病菌的产生,从而改变疾病的发展。 Abstract:Objective To investigate the composition and functional change of intestinal microbiota in infantile cholestasis (IC). Methods A total of 42 infants with IC who were admitted to Shenzhen Children's Hospital from September 2014 to February 2016 were enrolled as IC group, and 32 normal infants during the same period of time were enrolled as control group. Fecal samples were collected, and shotgun sequencing was used to investigate the features of intestinal microbiota. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and eggNOG database were used to investigate the functional changes of intestinal microbiota. The Wilcoxon rank-sum test was used for comparison of continuous data between groups, and the chi-square test was used for comparison of categorical data between groups. Shannon coefficient, the Wilcoxon Rank sum test, and DESeq2 were used to calculate and analyze the data of strains and the functional pathways of genes, and Cytoscape was used to give a visual representation of abundant strains and their association. Results As for the etiology of IC in the IC group, 11(26.2%) had biliary atresia, 4(9.5%) had Citrin deficiency, 4(9.5%) had bile duct dysplasia, and 23(54.8%) had unknown cause. There was a significant difference in the composition of intestinal microbiota at the phylum level between the two groups (Z=0.000 046, P < 0.05), and the IC group had significant increases in Firmicutes and Bacteroidetes. There were also significant differences between the two groups at the species level (Z=0.000 014, P < 0.05), and the IC group had significant reductions in the probiotic bacteria including Bifidobacterium and significant increases in the potential pathogens including Veillonella and Niastella koreensis. There was a significant difference in the function of intestinal microbiota between the two groups (Z=0.019 8, P < 0.05), and the IC group had significant reductions in the functions of intestinal microbiota such as amino acid metabolism and carbohydrate metabolism. Conclusion Changes in the composition and function of intestinal microbiota are observed in infants with IC, with a significant reduction in probiotic bacteria and a significant increase in potential pathogens, and supplementation with probiotic bacteria including Bifidobacterium can reduce the production of potential pathogens and change disease progression. -
Key words:
- Infant /
- Cholestasis /
- Gastrointestinal Microbiome
-
图 3 胆汁淤积组和对照组中较丰富的菌种交互网络图
注:由Cytoscape展示的交互网络图,农研丝杆菌GR20-10(Niastella koreensis GR20-10)、韦永氏球菌DSM 2008(Veillonella parvula DSM 2008)、加氏乳杆菌ATCC33323(Lactobacillus gasseri ATCC 33323)和沼泽红假单胞菌BisB18(Rhodopseudomonas palustris BisB18)在胆汁淤积组中极为丰富。双歧杆菌(Bifidobacterium)如长双歧杆菌(Bifidobacterium longum)、齿双歧杆菌Bd1(Bifidobacterium dentium Bd1)和短双歧杆菌ACS-071-V-Sch8b(Bifidobacterium breve ACS-071-V-Sch8b)在对照组中极为丰富,狄氏副拟杆菌ATCC 8503(Parabacteroides distasonis ATCC 8503)在对照组中也是较丰富的。
表 1 2组患儿一般资料比较
项目 胆汁淤积组(n=42) 对照组(n=32) 统计值 P值 男/女(例) 21/21 25/7 χ2=6.11 0.013 就诊年龄(d) 90.0(29.0~300.0) 60.0(30.0~120.0) Z=4.88 0.019 分娩方式[例(%)] χ2=0.03 0.873 顺产 36(85.7) 27(84.4) 剖腹产 6(14.3) 5(15.6) 喂养方式[例(%)] χ2=2.55 0.111 母乳喂养 34(81.0) 30(93.8) 人工喂养 8(19.0) 2(6.2) TBil(μmol/L) 172.2(41.4~348.9) DBil(μmol/L) 78.3(21.4~153.9) IBil(μmol/L) 80.4(20.0~241.1) 胆道闭锁[例(%)] 11(26.2) Citrin蛋白缺乏症[例(%)] 4(9.5) 胆管发育不良[例(%)] 4(9.5) 原因未明[例(%)] 23(54.8) -
[1] LIU Y, HUANG ZH. Clinical diagnosis and treatment of cholestatic hepatopathy in infants[J]. J Clin Hepatol, 2015, 31(8): 1218-1220. (in Chinese) DOI: 10.3969/j.issn.1001-5256.2015.08.009刘艳, 黄志华. 婴儿胆汁淤积性肝病的诊断与治疗[J]. 临床肝胆病杂志, 2015, 31(8): 1218-1220. DOI: 10.3969/j.issn.1001-5256.2015.08.009 [2] FISCHLER B, LAMIREAU T. Cholestasis in the newborn and infant[J]. Clin Res Hepatol Gastroenterol, 2014, 38(3): 263-267. DOI: 10.1016/j.clinre.2014.03.010 [3] CESARO C, TISO A, DEL PRETE A, et al. Gut microbiota and probiotics in chronic liver diseases[J]. Dig Liver Dis, 2011, 43(6): 431-438. DOI: 10.1016/j.dld.2010.10.015 [4] TRAUNER M, FICKERT P, TILG H. Bile acids as modulators of gut microbiota linking dietary habits and inflammatory bowel disease: A potentially dangerous liaison[J]. Gastroenterology, 2013, 144(4): 844-846. DOI: 10.1053/j.gastro.2013.02.029 [5] TRAUNER M, HALILBASIC E. Nuclear receptors as new perspective for the management of liver diseases[J]. Gastroenterology, 2011, 140(4): 1120-1125. e1-e12. DOI: 10.1053/j.gastro.2011.02.044 [6] QIN N, YANG F, LI A, et al. Alterations of the human gut microbiome in liver cirrhosis[J]. Nature, 2014, 513(7516): 59-64. DOI: 10.1038/nature13568 [7] ZHOU S, XU R, HE F, et al. Diversity of gut microbiota metabolic pathways in 10 pairs of Chinese infant twins[J]. PLoS One, 2016, 11(9): e0161627. DOI: 10.1371/journal.pone.0161627 [8] HUSON DH, AUCH AF, QI J, et al. MEGAN analysis of metagenomic data[J]. Genome Res, 2007, 17(3): 377-386. DOI: 10.1101/gr.5969107 [9] DIXON P. VEGAN, a package of R functions for community ecology[J]. J Veg Sci, 2003, 14(6): 927-930. DOI: 10.1111/j.1654-1103.2003.tb02228.x [10] MANYAM G, BIRERDINC A, BARANOVA A. KPP: KEGG pathway painter[J]. BMC Syst Biol, 2015, 9(Suppl 2): s3. DOI: 10.1186/1752-0509-9-S2-S3 [11] JENSEN LJ, JULIEN P, KUHN M, et al. Eggnog: Automated construction and annotation of orthologous groups of genes[J]. Nucleic Acids Res, 2008, 36(Suppl 1): d250-d254. http://europepmc.org/abstract/MED/17942413 [12] LOVE MI, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. DOI: 10.1186/s13059-014-0550-8 [13] SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13(11): 2498-2504. DOI: 10.1101/gr.1239303 [14] HUANG Q, ZHANG HB, LI JT, et al. Research advances in the mechanism of action of intestinal microecology in intrahepatic cholestasis[J]. J Clin Hepatol, 2019, 35(10): 2355-2359. (in Chinese) DOI: 10.3969/j.issn.1001-5256.2019.10.050黄倩, 张海博, 李京涛, 等. 肠道微生态在肝内胆汁淤积中的作用机制[J]. 临床肝胆病杂志, 2019, 35(10): 2355-2359. DOI: 10.3969/j.issn.1001-5256.2019.10.050 [15] RIDLON JM, ALVES JM, HYLEMON PB, et al. Cirrhosis, bile acids and gut microbiota: Unraveling a complex relationship[J]. Gut Microbes, 2013, 4(5): 382-387. DOI: 10.4161/gmic.25723 [16] COSTELLO EK, STAGAMAN K, DETHLEFSEN L, et al. The application of ecological theory toward an understanding of the human microbiome[J]. Science, 2012, 336(6086): 1255-1262. DOI: 10.1126/science.1224203 [17] TROSVIK P, STENSETH NC, RUDI K. Convergent temporal dynamics of the human infant gut microbiota[J]. ISME J, 2010, 4(2): 151-158. DOI: 10.1038/ismej.2009.96 [18] KOENIG JE, SPOR A, SCALFONE N, et al. Succession of microbial consortia in the developing infant gut microbiome[J]. Proc Natl Acad Sci U S A, 2011, 108 (Suppl 1): 4578-4585. http://emph.oxfordjournals.org/cgi/ijlink?linkType=ABST&journalCode=pnas&resid=108/Supplement_1/4578 [19] OLSZAK T, AN D, ZEISSIG S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function[J]. Science, 2012, 336(6080): 489-493. DOI: 10.1126/science.1219328 [20] CHEN Y, YANG F, LU H, et al. Characterization of fecal microbial communities in patients with liver cirrhosis[J]. Hepatology, 2011, 54(2): 562-572. DOI: 10.1002/hep.24423 [21] YANG L, GE WP, WANG R, et al. Influence of age, gender and delivery mode on the differential analysis of intestinal flora in infants[J]. Acta Nutrimenta Sinica, 2019, 41(4): 352-357. (in Chinese) https://www.cnki.com.cn/Article/CJFDTOTAL-YYXX201904015.htm杨莉, 葛武鹏, 王瑞, 等. 不同月龄、性别、分娩方式对婴儿肠道菌群差异性分析[J]. 营养学报, 2019, 41(4): 352-357. https://www.cnki.com.cn/Article/CJFDTOTAL-YYXX201904015.htm [22] BÄCKHED F, ROSWALL J, PENG Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life[J]. Cell Host Microbe, 2015, 17(6): 852. DOI: 10.1016/j.chom.2015.05.012 [23] BAJAJ JS, RIDLON JM, HYLEMON PB, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy[J]. Am J Physiol Gastrointest Liver Physiol, 2012, 302(1): g168-g175. DOI: 10.1152/ajpgi.00190.2011 [24] FUKUI H. Gut-liver axis in liver cirrhosis: How to manage leaky gut and endotoxemia[J]. World J Hepatol, 2015, 7(3): 425-442. DOI: 10.4254/wjh.v7.i3.425 [25] BAUER TM, SCHWACHA H, STEINBRVCKNER B, et al. Small intestinal bacterial overgrowth in human cirrhosis is associated with systemic endotoxemia[J]. Am J Gastroenterol, 2002, 97(9): 2364-2370. DOI: 10.1111/j.1572-0241.2002.05791.x [26] WANG Y, GAO X, ZHANG X, et al. Gut microbiota dysbiosis is associated with altered bile acid metabolism in infantile cholestasis[J]. mSystems, 2019, 4(6): e00463-19. http://www.ncbi.nlm.nih.gov/pubmed/31848302 [27] QIN J, LI Y, CAI Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes[J]. Nature, 2012, 490(7418): 55-60. DOI: 10.1038/nature11450 [28] GREENBLUM S, TURNBAUGH PJ, BORENSTEIN E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease[J]. Proc Natl Acad Sci U S A, 2012, 109(2): 594-599. DOI: 10.1073/pnas.1116053109 [29] LEPAGE P, HÄSLER R, SPEHLMANN ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis[J]. Gastroenterology, 2011, 141(1): 227-236. DOI: 10.1053/j.gastro.2011.04.011 -


 PDF下载 ( 4055 KB)
PDF下载 ( 4055 KB)

 下载:
下载: