Role of microRNAs in the development and progression of nonalcoholic steatohepatitis
-
摘要: 非酒精性脂肪性肝炎(NASH)已经成为肝炎的第二大病因,可进一步进展为肝纤维化、肝硬化甚至肝癌,但其确切发病机制尚不十分清楚,亦缺乏有效的治疗药物。microRNA是体内的一类非编码、转录后调控、高度保守的小分子RNA,在多种肝脏疾病中发挥着重要作用,主要就microRNA在NASH发生发展中的作用作一综述。Abstract: Nonalcoholic steatohepatitis (NASH) has become the second leading cause of hepatitis and can further progress to liver fibrosis, liver cirrhosis, and even liver cancer; however, the detailed pathogenesis of NASH remains unclear, and there is still a lack of effective therapeutic drugs. MicroRNAs (miRNAs) are a class of non-coding, post-transcriptionally regulated, and highly conserved small RNAs in the body and play an important role in a variety of liver diseases. This article mainly reviews the role of miRNAs in the development and progression of NASH.
-
Key words:
- Nonalcoholic Steatohepatitis /
- MicroRNAs /
- Pathologic Processes
-
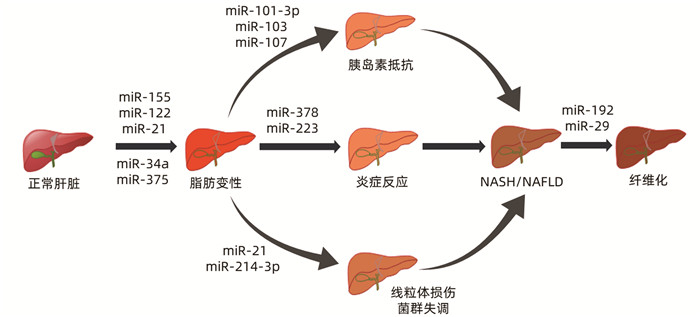
表 1 miRNA在NASH中的表达
miRNA 表达异常 作用机制 参考文献 miR-155 表达下调 结合LXRα的3′UTR促进肝脂肪生成;增强C/EBP的丰度 [8-11] miR-122 表达上调 通过上调SREBP-1c、HMG-CoA、SREBP-2影响脂质代谢, 参与线粒体氧化应激 [12-13] miR-21 表达上调 抑制PPARα的表达,导致脂质堆积;参与菌群失调的调节 [14-15] miR-375 表达上调 抑制PPARα通路的激活影响肝脏脂质代谢及炎症反应 [16] miR-34a 表达上调 引起其靶点PPARα和SIRT1的下调,从而导致脂质的异常沉积,激活AMPK途径促进炎症进展 [17-19] miR-103/107 表达上调 增加脂肪细胞中直接靶基因小窝蛋白1的表达,诱导胰岛素抵抗 [20] miR-101-3p 表达下调 通过调节胰岛素信号的关键介质来促进胰岛素抵抗 [21] miR-378 表达上调 阻断AMPK信号通路而增强NF-κB/TNFα轴,增强炎症反应 [22] miR-223 表达上调 抑制CXCL10和TAZ的表达,影响肝脏炎症 [23] miR-214-3p 表达上调 抑制线粒体自噬 [24] miR-192 表达上调 通过激活TGFβ/Smad信号,参与肝纤维化过程 [25] miR-29 表达下调 参与了胰岛素信号通路、Wnt信号通路、丝裂原活化蛋白激酶信号通路, 抑制UPRmt的激活 [26] 注:SREBP,固醇调节元件结合蛋白;HMG-CoA,3-羟基-3-甲基戊二酰辅酶A;PPAR,过氧化物酶体增殖物激活受体;SIRT,沉默信息调节因子;AMPK,AMP依赖的蛋白激酶;CXCL,趋化因子;TAZ,PDZ结合基序;LXR,肝脏X受体;UTR,非翻译区。 -
[1] LONG JL, XU H, ZENG M, et al. Research progress in diagnosis of non-alcoholic steatohepatitis[J]. Progr Pharm Sci, 2020, 44(3): 201-207. https://www.cnki.com.cn/Article/CJFDTOTAL-YXJZ202003007.htm龙健灵, 徐蕙, 曾梦, 等. 非酒精性脂肪性肝炎诊断研究进展[J]. 药学进展, 2020, 44(3): 201-207. https://www.cnki.com.cn/Article/CJFDTOTAL-YXJZ202003007.htm [2] CHI ZC. Pathogenesis of nonalcoholic steatohepatitis and cirrhosis[J]. J Pract Hepatol, 2018, 21(2): 166-169. DOI: 10.3969/j.issn.1672-5069.2018.02.005.池肇春. 非酒精性脂肪性肝炎肝硬化发病机制研究进展与展望[J]. 实用肝脏病杂志, 2018, 21(2): 166-169. DOI: 10.3969/j.issn.1672-5069.2018.02.005. [3] CALIGIURI A, GENTILINI A, MARRA F. Molecular pathogenesis of NASH[J]. Int J Mol Sci, 2016, 17(9): 1575. DOI: 10.3390/ijms17091575. [4] LU TX, ROTHENBERG ME. MicroRNA[J]. J Allergy Clin Immunol, 2018, 141(4): 1202-1207. DOI: 10.1016/j.jaci.2017.08.034. [5] SZABO G, CSAK T. Role of MicroRNAs in NAFLD/NASH[J]. Dig Dis Sci, 2016, 61(5): 1314-1324. DOI: 10.1007/s10620-015-4002-4. [6] PIROLA CJ, FERNÁNDEZ GIANOTTI T, CASTAÑO GO, et al. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis[J]. Gut, 2015, 64(5): 800-812. DOI: 10.1136/gutjnl-2014-306996. [7] CHEUNG O, PURI P, EICKEN C, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression[J]. Hepatology, 2008, 48(6): 1810-1820. DOI: 10.1002/hep.22569. [8] WANG L, ZHANG N, WANG Z, et al. Decreased MiR-155 level in the peripheral blood of non-alcoholic fatty liver disease patients may serve as a biomarker and may influence LXR activity[J]. Cell Physiol Biochem, 2016, 39(6): 2239-2248. DOI: 10.1159/000447917. [9] CHEN Y, SIEGEL F, KIPSCHULL S, et al. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit[J]. Nat Commun, 2013, 4: 1769. DOI: 10.1038/ncomms2742. [10] KAJIMURA S, SEALE P, SPIEGELMAN BM. Transcriptional control of brown fat development[J]. Cell Metab, 2010, 11(4): 257-262. DOI: 10.1016/j.cmet.2010.03.005. [11] HE M, XU Z, DING T, et al. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta[J]. Cell Mol Immunol, 2009, 6(5): 343-352. DOI: 10.1038/cmi.2009.45. [12] OTSUKA M, KISHIKAWA T, YOSHIKAWA T, et al. MicroRNAs and liver disease[J]. J Hum Genet, 2017, 62(1): 75-80. DOI: 10.1038/jhg.2016.53. [13] ZHANG R, WANG X, QU JH, et al. Caloric restriction induces microRNAs to improve mitochondrial proteostasis[J]. iScience, 2019, 17: 155-166. DOI: 10.1016/j.isci.2019.06.028. [14] LOYER X, PARADIS V, HÉNIQUE C, et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression[J]. Gut, 2016, 65(11): 1882-1894. DOI: 10.1136/gutjnl-2014-308883. [15] BLASCO-BAQUE V, COUPÉ B, FABRE A, et al. Associations between hepatic miRNA expression, liver triacylglycerols and gut microbiota during metabolic adaptation to high-fat diet in mice[J]. Diabetologia, 2017, 60(4): 690-700. DOI: 10.1007/s00125-017-4209-3. [16] LEI L, ZHOU C, YANG X, et al. Down-regulation of microRNA-375 regulates adipokines and inhibits inflammatory cytokines by targeting AdipoR2 in non-alcoholic fatty liver disease[J]. Clin Exp Pharmacol Physiol, 2018, 45(8): 819-831. DOI: 10.1111/1440-1681.12940. [17] DING J, LI M, WAN X, et al. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease[J]. Sci Rep, 2015, 5: 13729. DOI: 10.1038/srep13729. [18] CASTRO RE, FERREIRA DM, AFONSO MB, et al. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease[J]. J Hepatol, 2013, 58(1): 119-125. DOI: 10.1016/j.jhep.2012.08.008. [19] SIMÃO AL, AFONSO MB, RODRIGUES PM, et al. Skeletal muscle miR-34a/SIRT1: AMPK axis is activated in experimental and human non-alcoholic steatohepatitis[J]. J Mol Med (Berl), 2019, 97(8): 1113-1126. DOI: 10.1007/s00109-019-01796-8. [20] XU Q, LI Y, SHANG YF, et al. miRNA-103: Molecular link between insulin resistance and nonalcoholic fatty liver disease[J]. World J Gastroenterol, 2015, 21(2): 511-516. DOI: 10.3748/wjg.v21.i2.511. [21] MERONI M, LONGO M, ERCONI V, et al. mir-101-3p downregulation promotes fibrogenesis by facilitating hepatic stellate cell transdifferentiation during insulin resistance[J]. Nutrients, 2019, 11(11): 2597. DOI: 10.3390/nu11112597. [22] ZHANG T, HU J, WANG X, et al. MicroRNA-378 promotes hepatic inflammation and fibrosis via modulation of the NF-κB-TNFα pathway[J]. J Hepatol, 2019, 70(1): 87-96. DOI: 10.1016/j.jhep.2018.08.026. [23] HE Y, HWANG S, CAI Y, et al. MicroRNA-223 ameliorates nonalcoholic steatohepatitis and cancer by targeting multiple inflammatory and oncogenic genes in hepatocytes[J]. Hepatology, 2019, 70(4): 1150-1167. DOI: 10.1002/hep.30645. [24] HAMMOUTENE A, BIQUARD L, LASSELIN J, et al. A defect in endothelial autophagy occurs in patients with non-alcoholic steatohepatitis and promotes inflammation and fibrosis[J]. J Hepatol, 2020, 72(3): 528-538. DOI: 10.1016/j.jhep.2019.10.028. [25] COLL M, EL TAGHDOUINI A, PEREA L, et al. Integrative miRNA and gene expression profiling analysis of human quiescent hepatic stellate cells[J]. Sci Rep, 2015, 5: 11549. DOI: 10.1038/srep11549. [26] OGAWA T, ENOMOTO M, FUJⅡ H, et al. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis[J]. Gut, 2012, 61(11): 1600-1609. DOI: 10.1136/gutjnl-2011-300717. [27] RADHAKRISHNAN A, GOLDSTEIN JL, MCDONALD JG, et al. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance[J]. Cell Metab, 2008, 8(6): 512-521. DOI: 10.1016/j.cmet.2008.10.008. [28] LAMBERT G, AMAR MJ, GUO G, et al. The farnesoid X-receptor is an essential regulator of cholesterol homeostasis[J]. J Biol Chem, 2003, 278(4): 2563-2570. DOI: 10.1074/jbc.M209525200. [29] PAWLAK M, LEFEBVRE P, STAELS B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease[J]. J Hepatol, 2015, 62(3): 720-733. DOI: 10.1016/j.jhep.2014.10.039. [30] LI CM, WEI YH, LYU JY, et al. Research progress of Sirt1 and NAFLD pathogenesis and related drugs[J]. Med Recapitulate, 2018, 24(2): 214-219, 225. DOI: 10.3969/j.issn.1006-2084.2018.02.002.李春媚, 魏云华, 吕俊衍, 等. Sirt1与NAFLD发病机制及药物研究进展[J]. 医学综述, 2018, 24(2): 214-219, 225. DOI: 10.3969/j.issn.1006-2084.2018.02.002. [31] DUCHEIX S, MONTAGNER A, THEODOROU V, et al. The liver X receptor: A master regulator of the gut-liver axis and a target for non alcoholic fatty liver disease[J]. Biochem Pharmacol, 2013, 86(1): 96-105. DOI: 10.1016/j.bcp.2013.03.016. [32] KORACH-ANDRÉ M, GUSTAFSSON JÅ. Liver X receptors as regulators of metabolism[J]. Biomol Concepts, 2015, 6(3): 177-190. DOI: 10.1515/bmc-2015-0007. [33] SALMINEN A, HYTTINEN JM, KAARNIRANTA K. AMP-activated protein kinase inhibits NF-κB signaling and inflammation: Impact on healthspan and lifespan[J]. J Mol Med (Berl), 2011, 89(7): 667-676. DOI: 10.1007/s00109-011-0748-0. [34] SPAHIS S, DELVIN E, BORYS JM, et al. Oxidative stress as a critical factor in nonalcoholic fatty liver disease pathogenesis[J]. Antioxid Redox Signal, 2017, 26(10): 519-541. DOI: 10.1089/ars.2016.6776. [35] YANG YL, WANG PW, WANG FS, et al. miR-29a modulates GSK3β/SIRT1-linked mitochondrial proteostatic stress to ameliorate mouse non-alcoholic steatohepatitis[J]. Int J Mol Sci, 2020, 21(18): 6884. DOI: 10.3390/ijms21186884. [36] MARRA F, SVEGLIATI-BARONI G. Lipotoxicity and the gut-liver axis in NASH pathogenesis[J]. J Hepatol, 2018, 68(2): 280-295. DOI: 10.1016/j.jhep.2017.11.014. [37] KWIECINSKI M, NOETEL A, ELFIMOVA N, et al. Hepatocyte growth factor (HGF) inhibits collagen I and IV synthesis in hepatic stellate cells by miRNA-29 induction[J]. PLoS One, 2011, 6(9): e24568. DOI: 10.1371/journal.pone.0024568. [38] van der REE MH, de VREE JM, STELMA F, et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: A phase 1B, double-blind, randomised controlled trial[J]. Lancet, 2017, 389(10070): 709-717. DOI: 10.1016/S0140-6736(16)31715-9. [39] JANSSEN HL, REESINK HW, LAWITZ EJ, et al. Treatment of HCV infection by targeting microRNA[J]. N Engl J Med, 2013, 368(18): 1685-1694. DOI: 10.1056/NEJMoa1209026. -


 PDF下载 ( 2298 KB)
PDF下载 ( 2298 KB)

 下载:
下载:


