miR-409-3p在肝癌HepG2细胞中的表达及其在细胞增殖中的作用机制
DOI: 10.3969/j.issn.1001-5256.2023.08.019
Expression of miR-409-3p in hepatoma HepG2 cells and its mechanism in cell proliferation
-
摘要:
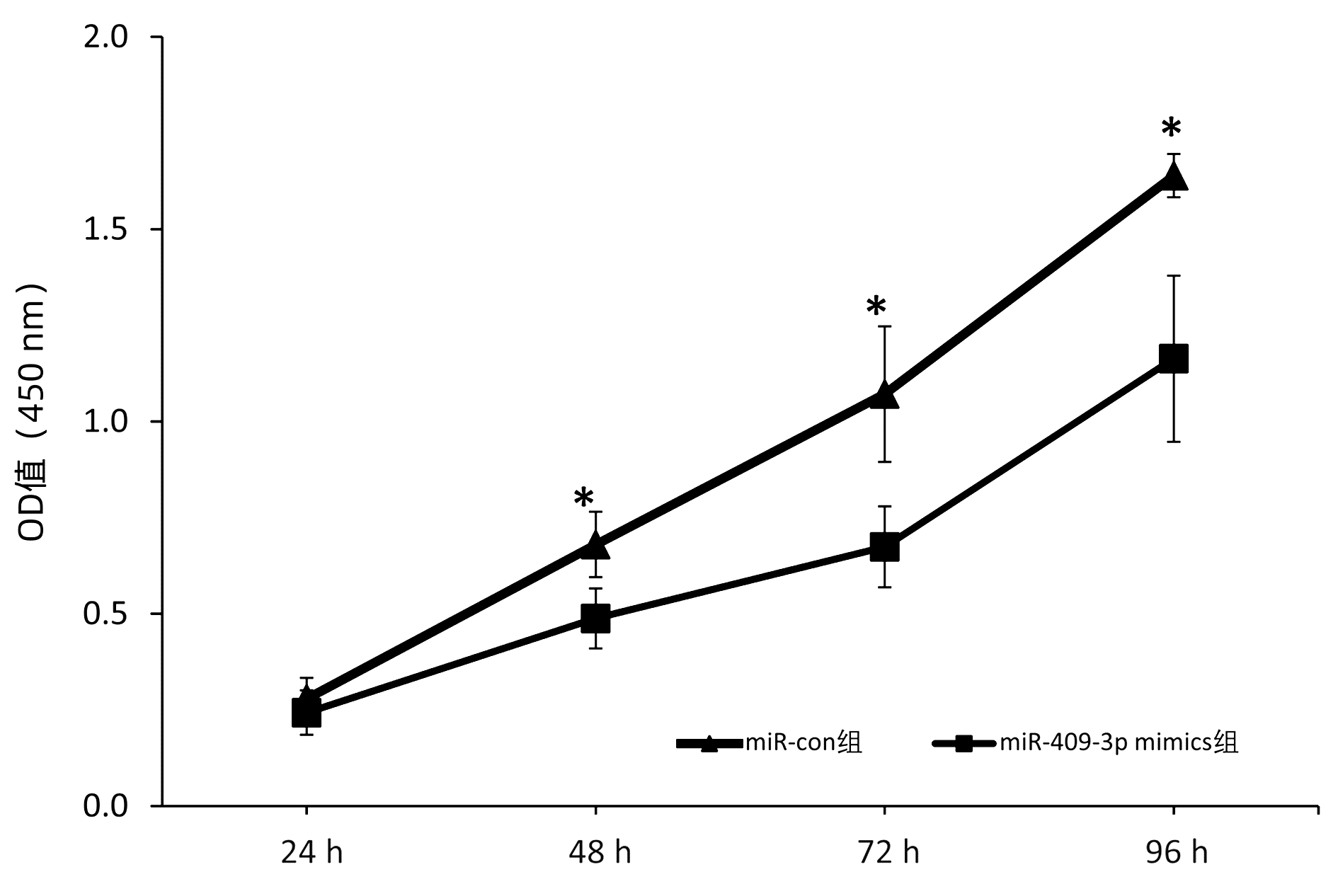
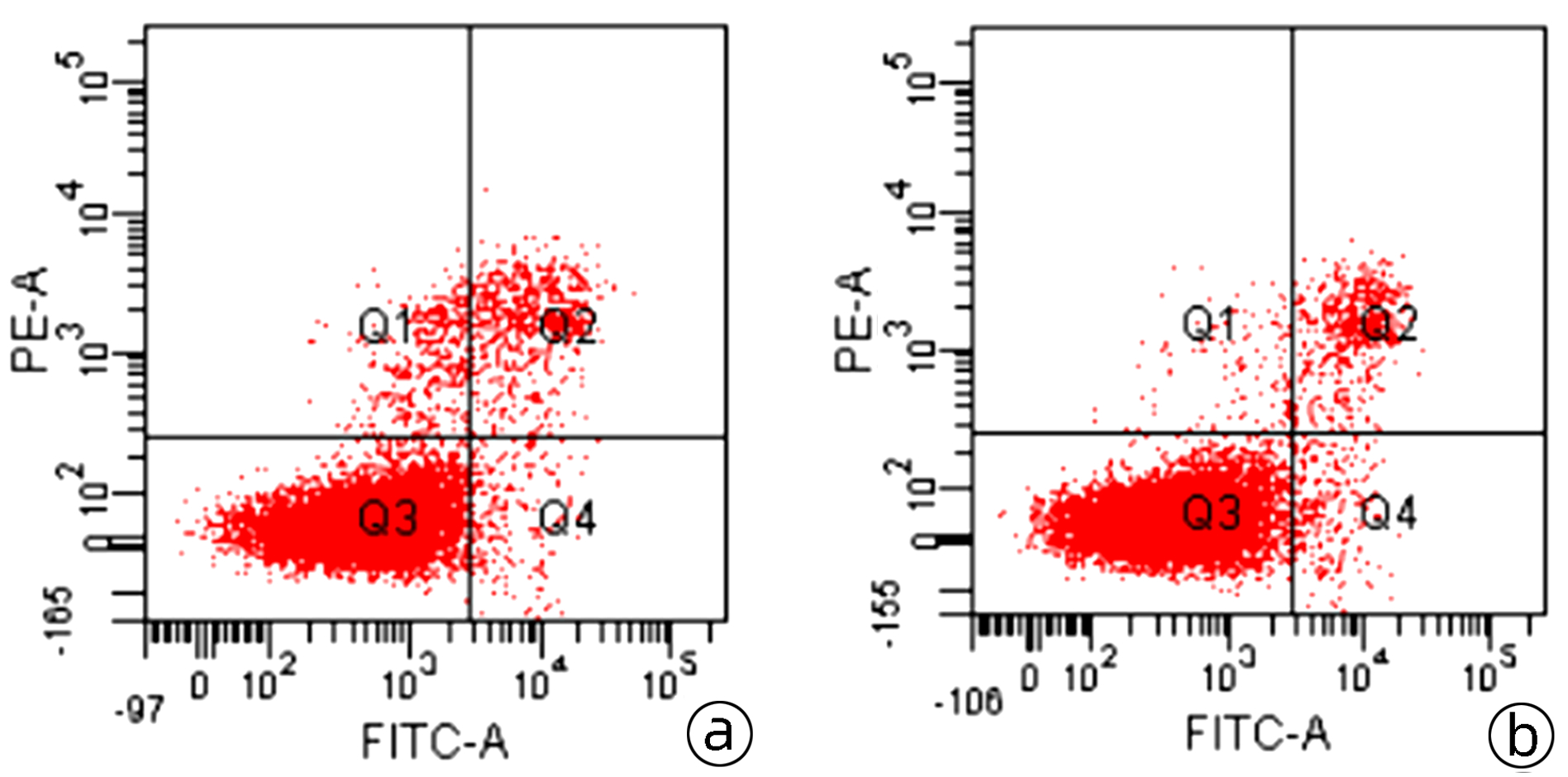
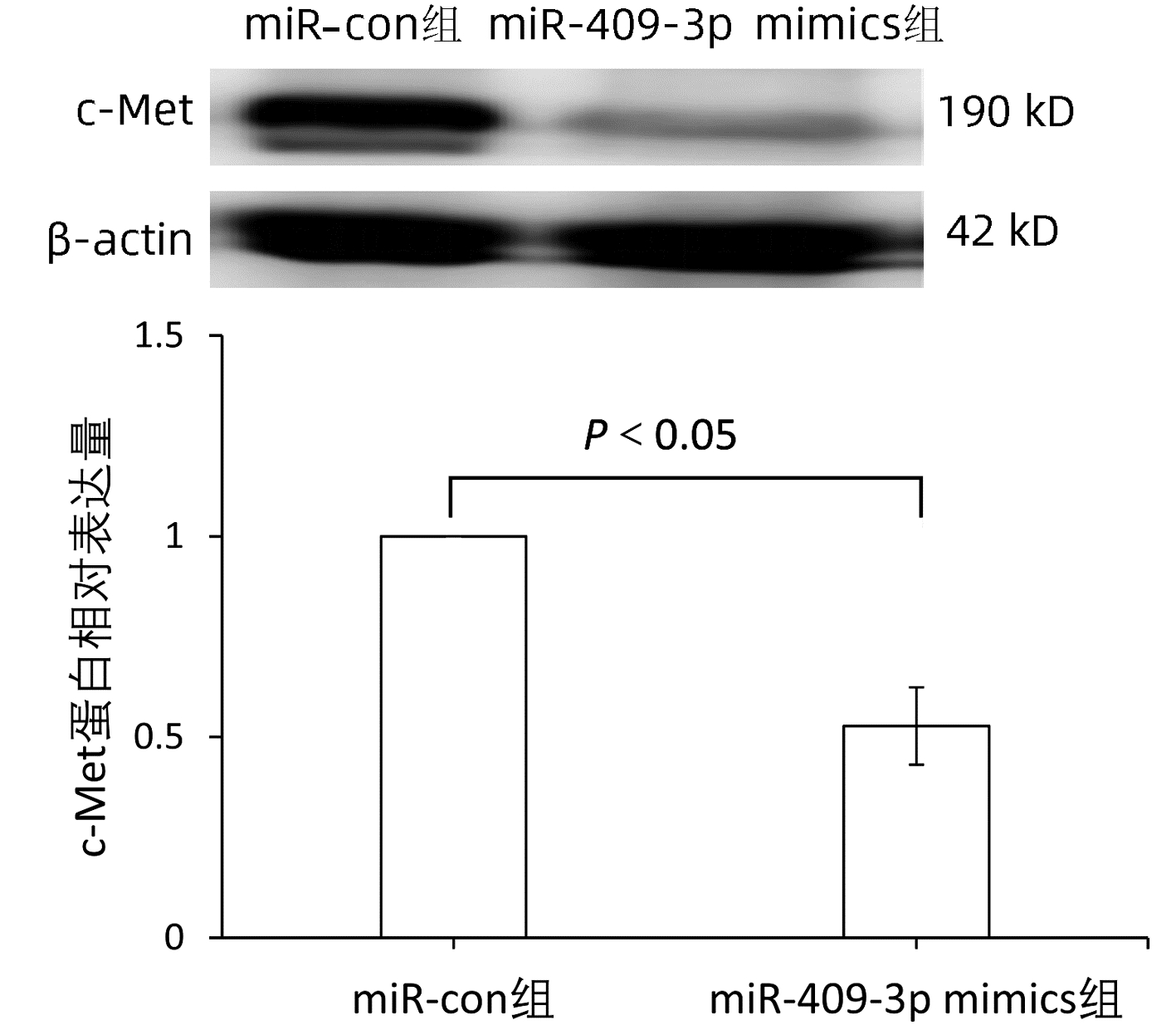
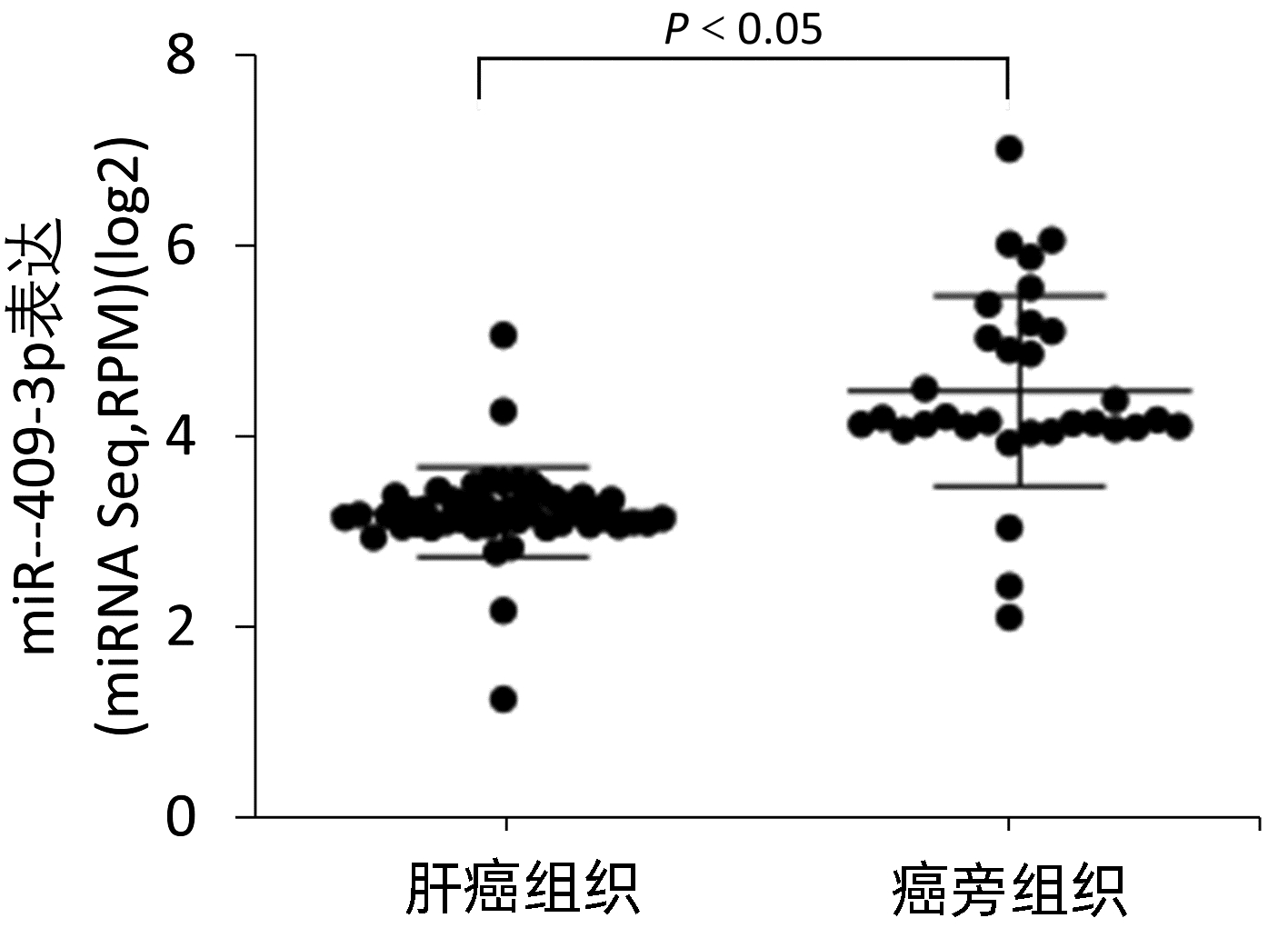
目的 本研究旨在明确miR-409-3p在肝癌细胞系中的表达及其意义,并探讨可能的分子机制。 方法 利用实时荧光定量PCR的方法检测miR-409-3p在正常肝细胞LO2,以及HepG2、BEL-7402、SMMC-7721、MHCC-97H四种肝癌细胞中的表达差异。阳离子脂质体法将miR-409-3p mimics及microRNA mimics control瞬时转染至肝癌细胞株HepG2中,利用CCK8法、平板克隆、流式细胞术检测miR-409-3p对体外癌细胞增殖、细胞周期及凋亡的影响。Western Blot检测过表达miR-409-3p的HepG2细胞中c-Met蛋白的表达变化,荧光素酶报告系统鉴定靶向关系。计量资料两组间比较采用成组t检验,多组间比较采用单因素方差分析,进一步两两比较采用SNK法。 结果 基于TCGA肝癌microRNA表达谱数据,肝癌组织中miR-409-3p的表达水平显著低于癌旁组织(t=7.752,P<0.05)。与在LO2中的表达水平相比,miR-409-3p在HepG2、SMMC-7721、MHCC-97H、BEL-7402中的表达水平均明显降低(F=31.043,P<0.05)。与miR-con组相比,转染miR-409-3p mimics后的HepG2细胞中miR-409-3p的表达水平上升(t=-8.836,P<0.05),说明干扰有效。CCK8实验结果显示,与miR-con组相比,转染miR-409-3p mimics后的HepG2细胞增殖能力在48、72、96 h明显减弱,差异均具有统计学意义(t值分别为2.876、3.359、3.707,P值均<0.05)。平板克隆形成实验显示,miR-409-3p mimics组的细胞克隆形成率明显低于miR-con组(t=2.846,P=0.047)。流式细胞术结果显示,与miR-con组相比,过表达miR-409-3p后导致HepG2细胞G2期细胞数增多,差异具有统计学意义(t=-3.763,P<0.05);而凋亡率无明显统计学差异(t=0.714,P=0.515)。荧光素酶报告系统鉴定结果显示c-Met为miR-409-3p的靶基因(t=4.970,P=0.007)。与miR-con组相比,转染miR-409-3p mimics的HepG2细胞中,c-Met蛋白表达水平下降(t=-8.509,P=0.001)。 结论 miR-409-3p通过抑制c-Met蛋白表达,进而调控下游信号通路引起细胞周期G2期阻滞,从而抑制肝癌HepG2细胞的增殖。 -
关键词:
- 癌, 肝细胞 /
- 微RNAs /
- 原癌基因蛋白质c-met /
- 细胞增殖
Abstract:Objective To investigate the expression and significance of miR-409-3p in hepatoma carcinoma cell lines and possible molecular mechanism. Methods Quantitative real-time PCR was used to measure the expression of miR-409-3p in normal LO2 hepatocytes and four hepatoma cell line (HepG2, BEL-7402, SMMC-7721, and MHCC-97H). Hepatoma HepG2 cells were transiently transfected with miR-409-3p mimics and microRNA mimics control using the cationic liposome method, and then CCK8, plate colony formation assay, and flow cytometry were used to observe the effect of miR-409-3p on the proliferation, cell cycle, and apoptosis of hepatoma cells in vitro. Western blot was used to measure the change in the expression of c-Met protein in HepG2 cells with overexpression of miR-409-3p, and luciferase reporter gene assay was used to identify targeting relationship. The independent-samples t test was used for comparison between two groups, and a one-way analysis of variance was used for comparison between multiple groups, followed by the SNK test. Results Based on the TCGA microRNA expression profile data of liver cancer, the expression level of miR-409-3p in liver cancer tissue was significantly lower than that in adjacent tissue (t=7.752, P < 0.05). Compared with the LO2 cells, the HepG2, SMMC-7721, MHCC-97H, and BEL-7402 cells had a significant reduction in the expression level of miR-409-3p (F=31.043, P < 0.05). Compared with the miR-con group, the HepG2 cells transfected with miR-409-3p mimics had a significant increase in the expression level of miR-409-3p (t=-8.836, P < 0.05), suggesting that the interference was effective. CCK8 assay showed that compared with the miR-con group, the HepG2 cells transfected with miR-409-3p mimics had a significant reduction in proliferative capacity at 48, 72, and 96 hours (t=2.876, 3.359, and 3.707, all P < 0.05). Plate colony formation assay showed that the miR-409-3p mimics group had a significantly lower plating efficiency than the miR-con group (t=2.846, P=0.047). Flow cytometry showed that compared with the miR-con group, overexpression of miR-409-3p resulted in the increased number of HepG2 cells in G2 phase (t=-3.763, P < 0.05), while there was no significant difference in apoptosis rate (t=0.714, P=0.515). Luciferase reporter gene assay showed that c-Met was a target gene of miR-409-3p (t=4.970, P=0.007). Compared with the miR-con group, the HepG2 cells transfected with miR-409-3p mimics had a significant reduction in the expression of c-Met protein (t=-8.509, P=0.001). Conclusion By inhibiting the protein expression of c-Met, miR-409-3p regulates downstream signaling pathways to induce cell cycle arrest in G2 phase and thus inhibits the proliferation of hepatoma HepG2 cells. -
表 1 miR-409-3p在肝癌细胞及正常肝细胞中的表达
Table 1. Expression of miR-409-3p in hepatocellular carcinoma cells and normal hepatocyte
细胞种类 miR-409-3p相对表达量 LO2 1.000±0.000 HepG2 0.363±0.1261) BEL-7402 0.569±0.0951) SMMC-7721 0.838±0.0411) MHCC-97H 0.654±0.0511) F值 31.043 P值 <0.05 注:1)与LO2相比,P<0.05。 表 2 CCK8检测miR-409-3p转染后HepG2细胞体外增殖情况(OD值)
Table 2. In vitro proliferation of HepG2 cells after transfection with miR-409-3p detected by CCK8 (OD value)
时间 miR-con组 miR-409-3p mimics组 t值 P值 24 h 0.279±0.054 0.243±0.058 0.791 0.473 48 h 0.680±0.085 0.488±0.078 2.876 0.045 72 h 1.071±0.176 0.674±0.105 3.359 0.028 96 h 1.639±0.056 1.163±0.216 3.707 0.021 注:1)与24 h相比,P<0.05。 表 3 流式细胞仪检测miR-409-3p转染后HepG2细胞的各周期分布
Table 3. Cell cycle distribution of HepG2 cells after transfection with miR-409-3p detected by flow cytometry
细胞周期 miR con组(%) miR-409-3p mimics组(%) t值 P值 G1期 70.34±2.17 66.60±0.82 2.794 0.049 S期 23.55±0.35 22.54±1.09 1.534 0.200 G2期 6.11±1.81 10.86±1.22 -3.763 0.020 表 4 荧光素酶活性变化
Table 4. The change of luciferase activity
组别 MUT WT miR-con组 1.00±0.12 1.00±0.11 miR-409-3p mimics组 1.24±0.15 1.63±0.19 t值 2.254 4.970 P值 0.087 0.007 -
[1] RUIZ-MANRIQUEZ LM, CARRASCO-MORALES O, SANCHEZ Z EA, et al. MicroRNA-mediated regulation of key signaling pathways in hepatocellular carcinoma: A mechanistic insight[J]. Front Genet, 2022, 13: 910733. DOI: 10.3389/fgene.2022.910733. [2] COOPER CS, PARK M, BLAIR DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line[J]. Nature, 1984, 311(5981): 29-33. DOI: 10.1038/311029a0. [3] FAIELLA A, RICCARDI F, CARTENÌ G, et al. The emerging role of c-Met in carcinogenesis and clinical implications as a possible therapeutic target[J]. J Oncol, 2022, 2022: 5179182. DOI: 10.1155/2022/5179182. [4] CHEN L, SHI Y, ZHU X, et al. IL-10 secreted by cancer-associated macrophages regulates proliferation and invasion in gastric cancer cells via c-Met/STAT3 signaling[J]. Oncol Rep, 2019, 42(2): 595-604. DOI: 10.3892/or.2019.7206. [5] MA Y, ZHANG M, WANG J, et al. High-affinity human anti-c-Met IgG conjugated to oxaliplatin as targeted chemotherapy for hepatocellular carcinoma[J]. Front Oncol, 2019, 9: 717. DOI: 10.3389/fonc.2019.00717. [6] XU X, CHEN H, LIN Y, et al. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met[J]. Mol Cells, 2013, 36(1): 62-68. DOI: 10.1007/s10059-013-0044-7. [7] WAN L, ZHU L, XU J, et al. MicroRNA-409-3p functions as a tumor suppressor in human lung adenocarcinoma by targeting c-Met[J]. Cell Physiol Biochem, 2014, 34(4): 1273-1290. DOI: 10.1159/000366337. [8] General Office of National Health Commission. Standard for diagnosis and treatment of primary liver cancer (2022 edition)[J]. J Clin Hepatol, 2022, 38(2): 288-303. DOI: 10.3969/j.issn.1001-5256.2022.02.009.国家卫生健康委办公厅. 原发性肝癌诊疗指南(2022年版)[J]. 临床肝胆病杂志, 2022, 38(2): 288-303. DOI: 10.3969/j.issn.1001-5256.2022.02.009. [9] SIEGEL RL, MILLER KD, GODING SAUER A, et al. Colorectal cancer statistics, 2020[J]. CA Cancer J Clin, 2020, 70(3): 145-164. DOI: 10.3322/caac.21601. [10] FORNER A, LLOVET JM, BRUIX J. Hepatocellular carcinoma[J]. Lancet, 2012, 379(9822): 1245-1255. DOI: 10.1016/S0140-6736(11)61347-0. [11] RAY K. Liver cancer: The promise of new approaches in the management of hepatocellular carcinoma--adding to the toolbox?[J]. Nat Rev Gastroenterol Hepatol, 2013, 10(4): 195. DOI: 10.1038/nrgastro.2013.52. [12] TRINCHET JC, CHAFFAUT C, BOURCIER V, et al. Ultrasonographic surveillance of hepatocellular carcinoma in cirrhosis: a randomized trial comparing 3- and 6-month periodicities[J]. Hepatology, 2011, 54(6): 1987-1997. DOI: 10.1002/hep.24545. [13] ZHOU Q, SHAO JG. Research progress of miRNA in HBV-related hepatocellular carcinoma[J]. J Nantong Univ(Med Sci), 2022, 42(3): 257-261. DOI: 10.16424/j.cnki.cn32-1807/r.2022.03.013.周倩, 邵建国. MiRNA在HBV相关肝癌中的研究进展[J]. 南通大学学报(医学版), 2022, 42(3): 257-261. DOI: 10.16424/j.cnki.cn32-1807/r.2022.03.013. [14] XIE HJ, RASHED N, NING Y, et al. Current status of research on circulating microRNAs as diagnostic markers for hepatocellular carcinoma[J]. J Clin Hepatol, 2021, 37(2): 448-451. DOI: 10.3969/j.issn.1001-5256.2021.02.042.谢惠君, Rashed Nasot, 宁勇, 等. 循环miRNA作为肝细胞癌标志物的研究现状[J]. 临床肝胆病杂志, 2021, 37(2): 448-451. DOI: 10.3969/j.issn.1001-5256.2021.02.042. [15] HUSSEN BM, HIDAYAT HJ, SALIHI A, et al. MicroRNA: A signature for cancer progression[J]. Biomed Pharmacother, 2021, 138: 111528. DOI: 10.1016/j.biopha.2021.111528. [16] PIEROULI K, PAPAKONSTANTINOU E, PAPAGEORGIOU L, et al. Long non-coding RNAs and microRNAs as regulators of stress in cancer (Review)[J]. Mol Med Rep, 2022, 26(6): 361. DOI: 10.3892/mmr.2022.12878. [17] HUANG S, HE X. The role of microRNAs in liver cancer progression[J]. Br J Cancer, 2011, 104(2): 235-240. DOI: 10.1038/sj.bjc.6606010. [18] LIU S, LI B, XU J, et al. SOD1 Promotes cell proliferation and metastasis in non-small cell lung cancer via an miR-409-3p/SOD1/SETDB1 epigenetic regulatory feedforward loop[J]. Front Cell Dev Biol, 2020, 8: 213. DOI: 10.3389/fcell.2020.00213. [19] WANG Y, ZHANG J, CHEN X, et al. Circ_0001023 promotes proliferation and metastasis of gastric cancer cells thro ugh miR-409-3p/PHF10 axis[J]. Onco Targets Ther, 2020, 13: 4533-4544. DOI: 10.2147/OTT.S244358. [20] CUI X, CHEN J, ZHENG Y, et al. Circ_0000745 promotes the progression of cervical cancer by regulating miR-409-3p/ATF1 axis[J]. Cancer Biother Radiopharm, 2022, 37(9): 766-778. DOI: 10.1089/cbr.2019.3392. [21] CHEN J, WANG R, LU E, et al. LINC00630 as a miR-409-3p sponge promotes apoptosis and glycolysis of colon carcinoma cells via regulating HK2[J]. Am J Transl Res, 2022, 14(2): 863-875. [22] YANG S, ZOU C, LI Y, et al. Knockdown circTRIM28 enhances tamoxifen sensitivity via the miR-409-3p/HMGA2 axis in breast cancer[J]. Reprod Biol Endocrinol, 2022, 20(1): 146. DOI: 10.1186/s12958-022-01011-3. [23] KOTA J, CHIVUKULA RR, O'DONNELL KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model[J]. Cell, 2009, 137(6): 1005-1017. DOI: 10.1016/j.cell.2009.04.021. [24] XU T, ZHU Y, XIONG Y, et al. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells[J]. Hepatology, 2009, 50(1): 113-121. DOI: 10.1002/hep.22919. [25] XIAO F, ZHANG W, CHEN L, et al. MicroRNA-503 inhibits the G1/S transition by downregulating cyclin D3 and E2F3 in hepatocellular carcinoma[J]. J Transl Med, 2013, 11: 195. DOI: 10.1186/1479-5876-11-195. [26] FORNARI F, GRAMANTIERI L, FERRACIN M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma[J]. Oncogene, 2008, 27(43): 5651-5661. DOI: 10.1038/onc.2008.178. [27] LIU RF, XU X, HUANG J, et al. Down-regulation of miR-517a and miR-517c promotes proliferation of hepatocellular carcinoma cells via targeting Pyk2[J]. Cancer Lett, 2013, 329(2): 164-173. DOI: 10.1016/j.canlet.2012.10.027. [28] UEKI T, FUJIMOTO J, SUZUKI T, et al. Expression of hepatocyte growth factor and its receptor, the c-met proto-oncogene, in hepatocellular carcinoma[J]. Hepatology, 1997, 25(3): 619-623. DOI: 10.1002/hep.510250321. [29] GIORDANO S, COLUMBANO A. Met as a therapeutic target in HCC: facts and hopes[J]. J Hepatol, 2014, 60(2): 442-452. DOI: 10.1016/j.jhep.2013.09.009. [30] WANG Y, TAI Q, ZHANG J, et al. MiRNA-206 inhibits hepatocellular carcinoma cell proliferation and migration but promotes apoptosis by modulating cMET expression[J]. Acta Biochim Biophys Sin (Shanghai), 2019, 51(3): 243-253. DOI: 10.1093/abbs/gmy119. [31] LIU Y, TAN J, OU S, et al. MicroRNA-101-3p suppresses proliferation and migration in hepatocellular carcinoma by targeting the HGF/c-Met pathway[J]. Invest New Drugs, 2020, 38(1): 60-69. DOI: 10.1007/s10637-019-00766-8. [32] XU X, JIANG W, HAN P, et al. MicroRNA-128-3p Mediates Lenvatinib Resistance of Hepatocellular Carcinoma Cells by Downregulating c-Met[J]. J Hepatocell Carcinoma, 2022, 9: 113-126. DOI: 10.2147/JHC.S349369. [33] HUYNH H, ONG R, SOO KC. Foretinib demonstrates anti-tumor activity and improves overall survival in preclinical models of hepatocellular carcinoma[J]. Angiogenesis, 2012, 15(1): 59-70. DOI: 10.1007/s10456-011-9243-z. -


 PDF下载 ( 2369 KB)
PDF下载 ( 2369 KB)

 下载:
下载: