乙酰肝素酶在肝胆胰肿瘤中的作用及机制
DOI: 10.12449/JCH251136
-
摘要: 肝胆胰肿瘤是消化系统常见的疾病类型,由于其发病机制错综复杂,早期诊断困难且易转移,在我国发病率和致死率居高不下,严重影响患者生存质量和预后。研究显示,乙酰肝素酶作为基质重塑的关键效应分子,在肿瘤侵袭转移、微环境重塑中发挥关键调控作用,且与临床预后存在显著相关性。本文分析乙酰肝素酶在肝胆胰肿瘤中的分子作用机制,旨在为肝胆胰肿瘤的诊断标志物开发和靶向干预提供有力的科学依据。Abstract: Hepatobiliary and pancreatic tumors are common types of digestive system disorders, and their complex pathogeneses have led to difficulties in early diagnosis and a high metastasis rate, with persistently high incidence and mortality rates in China, which greatly affects the quality of life and prognosis of patients. Studies have shown that heparanase (HPSE), as a key effector molecule in matrix remodeling, plays a crucial regulatory role in tumor invasion and metastasis and microenvironment remodeling, and it is significantly associated with clinical prognosis. This article reviews the molecular mechanism of HPSE in hepatobiliary and pancreatic tumors, in order to provide a strong scientific basis for developing diagnostic markers and targeted interventions for hepatobiliary and pancreatic tumors.
-
Key words:
- Heparanase /
- Carcinoma, Hepatocellular /
- Cholangiocarcinoma /
- Pancreatic Neoplasms
-
表 1 HPSE与HPSE2的功能及分子机制对比
Table 1. Comparative of the functions and molecular mechanisms between HPSE and HPSE2
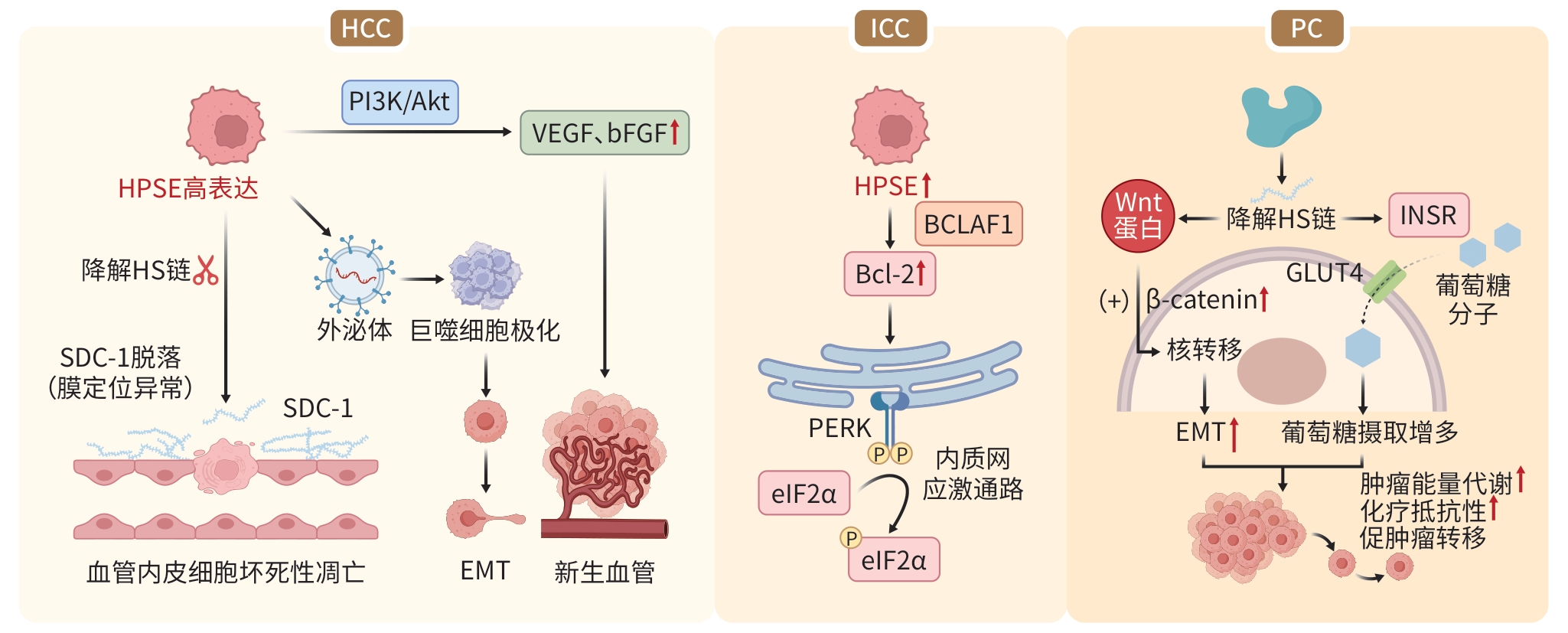
功能及分子机制 HPSE HPSE2 功能基础 ①具有HS降解活性,可裂解ECM及细胞表面的
HSPG[9];
②非酶活性参与调控外泌体形成、信号传导和基因转录缺乏HS降解活性,但对HS的亲和力高于HPSE,
能通过紧密结合HS抑制HPSE酶活性ECM降解 降解HS链,破坏基底膜结构,促进ECM降解[18] 通过C端肝素结合结构域与ECM及细胞表面的
HSPG高亲和力结合,阻碍HPSE与HS的结合及
切割,从而抑制ECM降解调控炎症反应 通过降解HS,释放细胞因子、生长因子、趋化因子等
多种炎症因子,加速炎症微环境的形成[18]通过竞争性结合HS,抑制细胞因子、生长因子、趋化
因子等多种炎症因子释放调控肿瘤转移、
侵袭力①释放VEGF、bFGF等促血管生成因子,促进肿瘤
新生血管生成;
②通过激活SDC-1、TNF-α诱导血管内皮细胞坏死性
凋亡;
③HPSE介导的核SDC-1缺失增强组蛋白乙酰转移酶
活性,促进驱动侵袭性肿瘤表型的基因表达[19]①抑制HS结合因子VEGF、bFGF等释放和血管
生成;
②上调Sox2表达,进而抑制EMT,阻止肿瘤细胞间
质化并抑制肿瘤侵袭能力[20];
③通过调节血管表面HS-生长因子的相互作用,维持
血管完整性[21]调控细胞凋亡 上调抗凋亡蛋白Bcl-2的表达水平,进一步激活由
PERK/eIF2α介导的内质网应激通路[22]①通过ATF3介导的内质网应激通路导致生长停滞
和细胞凋亡[23];
②诱导强肿瘤抑制因子(如BRD7)的表达和核定位,
减弱Erk信号传导,诱导促凋亡蛋白Bax的表达[24]调控肿瘤微环境 ①HPSE的过表达可能通过增加CD4 T细胞上PD-1和
CTLA-4的表达促进肿瘤生长[25];
②HPSE的过表达可能通过增加IL-35和减少T细胞
亚群产生的IFN-γ损伤T细胞的抗肿瘤能力[25]HPSE2可能促进巨噬细胞向M2型极化,塑造肿瘤
微环境,进而促进肿瘤生长[24]注:VEGF,血管内皮生长因子;bFGF,碱性成纤维细胞生长因子;Sox2,SRY相关高迁移率族盒蛋白2;EMT,上皮-间充质转化;PERK,蛋白激酶R样内质网激酶;eIF2α,真核细胞起始因子;ATF3,转录激活因子3;BRD7,溴结构域包含蛋白7;Erk,胞外信号调节激酶;PD-1,程序性死亡受体1;CTLA-4,细胞毒性T细胞相关抗原4。
-
[1] MINAMI K, MORIMOTO H, MORIOKA H, et al. Pathogenic roles of heparan sulfate and its use as a biomarker in mucopolysaccharidoses[J]. Int J Mol Sci, 2022, 23( 19): 11724. DOI: 10.3390/ijms231911724. [2] LING JX, LI JL, KHAN A, et al. Is heparan sulfate a target for inhibition of RNA virus infection?[J]. Am J Physiol Cell Physiol, 2022, 322( 4): C605- C613. DOI: 10.1152/ajpcell.00028.2022. [3] ALSHEHRI MA, ALSHEHRI MM, ALBALAWI NN, et al. Heparan sulfate proteoglycans and their modification as promising anticancer targets in hepatocellular carcinoma[J]. Oncol Lett, 2021, 21( 2): 173. DOI: 10.3892/ol.2021.12434. [4] FOOTE CA, SOARES RN, RAMIREZ-PEREZ FI, et al. Endothelial glycocalyx[J]. Compr Physiol, 2022, 12( 4): 3781- 3811. DOI: 10.1002/cphy.c210029. [5] ZAHAVI T, SALMON-DIVON M, SALGADO R, et al. Heparanase: A potential marker of worse prognosis in estrogen receptor-positive breast cancer[J]. NPJ Breast Cancer, 2021, 7( 1): 67. DOI: 10.1038/s41523-021-00277-x. [6] FARIA-RAMOS I, POÇAS J, MARQUES C, et al. Heparan sulfate glycosaminoglycans:(un)expected allies in cancer clinical management[J]. Biomolecules, 2021, 11( 2): 136. DOI: 10.3390/biom11020136. [7] SAAD F, GADALLAH M, DAIF A, et al. Heparanase(HPSE) gene polymorphism(rs12503843) contributes as a risk factor for hepatocellular carcinoma(HCC): A pilot study among Egyptian patients[J]. J Genet Eng Biotechnol, 2021, 19( 1): 3. DOI: 10.1186/s43141-020-00106-x. [8] YUAN FY, YANG YY, ZHOU HQ, et al. Heparanase in cancer progression: Structure, substrate recognition and therapeutic potential[J]. Front Chem, 2022, 10: 926353. DOI: 10.3389/fchem.2022.926353. [9] BARTOLINI B, CARAVÀ E, CAON I, et al. Heparan sulfate in the tumor microenvironment[J]. Adv Exp Med Biol, 2020, 1245: 147- 161. DOI: 10.1007/978-3-030-40146-7_7. [10] KOGANTI R, SURYAWANSHI R, SHUKLA D. Heparanase, cell signaling, and viral infections[J]. Cell Mol Life Sci, 2020, 77( 24): 5059- 5077. DOI: 10.1007/s00018-020-03559-y. [11] MASOLA V, ZAZA G, GAMBARO G, et al. Role of heparanase in tumor progression: Molecular aspects and therapeutic options[J]. Semin Cancer Biol, 2020, 62: 86- 98. DOI: 10.1016/j.semcancer.2019.07.014. [12] MAYFOSH AJ, NGUYEN TK, HULETT MD. The heparanase regulatory network in health and disease[J]. Int J Mol Sci, 2021, 22( 20): 11096. DOI: 10.3390/ijms222011096. [13] VLODAVSKY I, KAYAL Y, HILWI M, et al. Heparanase-A single protein with multiple enzymatic and nonenzymatic functions[J]. Proteoglycan Res, 2023, 1( 3): e6. DOI: 10.1002/pgr2.6. [14] YANG YY, YUAN FY, ZHOU HQ, et al. Potential roles of heparanase in cancer therapy: Current trends and future direction[J]. J Cell Physiol, 2023, 238( 5): 896- 917. DOI: 10.1002/jcp.30995. [15] PAPE T, HUNKEMÖLLER AM, KÜMPERS P, et al. Targeting the“sweet spot” in septic shock- A perspective on the endothelial glycocalyx regulating proteins Heparanase-1 and-2[J]. Matrix Biol Plus, 2021, 12: 100095. DOI: 10.1016/j.mbplus.2021.100095. [16] LIU JJ, KNANI I, GROSS-COHEN M, et al. Role of heparanase 2(Hpa2) in gastric cancer[J]. Neoplasia, 2021, 23( 9): 966- 978. DOI: 10.1016/j.neo.2021.07.010. [17] HOPKINS J, VOLETY I, QATANANI F, et al. Heparanase 2 modulation inhibits HSV-2 replication by regulating heparan sulfate[J]. Viruses, 2024, 16( 12): 1832. DOI: 10.3390/v16121832. [18] KHANNA M, PARISH CR. Heparanase: Historical aspects and future perspectives[J]. Adv Exp Med Biol, 2020, 1221: 71- 96. DOI: 10.1007/978-3-030-34521-1_3. [19] VLODAVSKY I, ILAN N, SANDERSON RD. Forty years of basic and translational heparanase research[J]. Adv Exp Med Biol, 2020, 1221: 3- 59. DOI: 10.1007/978-3-030-34521-1_1. [20] GROSS-COHEN M, YANKU Y, KESSLER O, et al. Heparanase 2(Hpa2) attenuates tumor growth by inducing Sox2 expression[J]. Matrix Biol, 2021, 99: 58- 71. DOI: 10.1016/j.matbio.2021.05.001. [21] BECKER Y, HALLER H. Current understanding of heparanase 2 regulation, a non-heparanase[J]. Biochem Soc Trans, 2025, 53( 1): BST 2024‑1281. DOI: 10.1042/BST20241281. [22] KALONI D, DIEPSTRATEN ST, STRASSER A, et al. BCL-2 protein family: Attractive targets for cancer therapy[J]. Apoptosis, 2023, 28( 1-2): 20- 38. DOI: 10.1007/s10495-022-01780-7. [23] KNANI I, SINGH P, GROSS-COHEN M, et al. Induction of heparanase 2(Hpa2) expression by stress is mediated by ATF3[J]. Matrix Biol, 2022, 105: 17- 30. DOI: 10.1016/j.matbio.2021.11.001. [24] SOBOH S, VORONTSOVA A, FARHOUD M, et al. Tumor- and host-derived heparanase-2(Hpa2) attenuates tumorigenicity: Role of Hpa2 in macrophage polarization and BRD7 nuclear localization[J]. Cell Death Dis, 2024, 15( 12): 894. DOI: 10.1038/s41419-024-07262-9. [25] ZHANG GL, GUTTER-KAPON L, ILAN N, et al. Significance of host heparanase in promoting tumor growth and metastasis[J]. Matrix Biol, 2020, 93: 25- 42. DOI: 10.1016/j.matbio.2020.06.001. [26] SHAH M, SARKAR D. HCC-related lncRNAs: Roles and mechanisms[J]. Int J Mol Sci, 2024, 25( 1): 597. DOI: 10.3390/ijms25010597. [27] CHEN XP, CHENG B, DAI DF, et al. Heparanase induces necroptosis of microvascular endothelial cells to promote the metastasis of hepatocellular carcinoma[J]. Cell Death Discov, 2021, 7( 1): 33. DOI: 10.1038/s41420-021-00411-5. [28] RESZEGI A, TÁTRAI P, REGŐS E, et al. Syndecan-1 in liver pathophysiology[J]. Am J Physiol Cell Physiol, 2022, 323( 2): C289- C294. DOI: 10.1152/ajpcell.00039.2022. [29] ZHANG XL, ZHAO YL, LIU LR, et al. Syndecan-1: A novel diagnostic and therapeutic target in liver diseases[J]. Curr Drug Targets, 2023, 24( 15): 1155- 1165. DOI: 10.2174/0113894501250057231102061624. [30] YANG R, CHEN MM, ZHENG JY, et al. The role of heparin and glycocalyx in blood-brain barrier dysfunction[J]. Front Immunol, 2021, 12: 754141. DOI: 10.3389/fimmu.2021.754141. [31] ARDIZZONE A, BOVA V, CASILI G, et al. Role of basic fibroblast growth factor in cancer: Biological activity, targeted therapies, and prognostic value[J]. Cells, 2023, 12( 7): 1002. DOI: 10.3390/cells1207-1002. [32] GALLARD C, LEBSIR N, KHURSHEED H, et al. Heparanase-1 is upregulated by hepatitis C virus and favors its replication[J]. J Hepatol, 2022, 77( 1): 29- 41. DOI: 10.1016/j.jhep.2022.01.008. [33] FENG F, WANG LJ, LI JC, et al. Role of heparanase in ARDS through autophagy and exosome pathway(review)[J]. Front Pharmacol, 2023, 14: 1200782. DOI: 10.3389/fphar.2023.1200782. [34] DAVID G, ZIMMERMANN P. Heparanase involvement in exosome formation[J]. Adv Exp Med Biol, 2020, 1221: 285- 307. DOI: 10.1007/978-3-030-34521-1_10. [35] van der VLAG J, BUIJSERS B. Heparanase in kidney disease[J]. Adv Exp Med Biol, 2020, 1221: 647- 667. DOI: 10.1007/978-3-030-34521-1_26. [36] YANG CG, DOU RZ, WEI C, et al. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis[J]. Mol Ther, 2021, 29( 6): 2088- 2107. DOI: 10.1016/j.ymthe.2021.02.006. [37] BRAGAZZI MC, VENERE R, RIBICHINI E, et al. Intrahepatic cholangiocarcinoma: Evolving strategies in management and treatment[J]. Dig Liver Dis, 2024, 56( 3): 383- 393. DOI: 10.1016/j.dld.2023.08.052. [38] YUAN FY, ZHOU HQ, LIU CY, et al. Heparanase interacting BCLAF1 to promote the development and drug resistance of ICC through the PERK/eIF2α pathway[J]. Cancer Gene Ther, 2024, 31( 6): 904- 916. DOI: 10.1038/s41417-024-00754-y. [39] HALBROOK CJ, LYSSIOTIS CA, PASCA DI MAGLIANO M, et al. Pancreatic cancer: Advances and challenges[J]. Cell, 2023, 186( 8): 1729- 1754. DOI: 10.1016/j.cell.2023.02.014. [40] XUE WH, YANG L, CHEN CX, et al. Wnt/β-catenin-driven EMT regulation in human cancers[J]. Cell Mol Life Sci, 2024, 81( 1): 79. DOI: 10.1007/s00018-023-05099-7. [41] WANG C, WEI YJ, WANG G, et al. Heparanase potentiates the invasion and migration of pancreatic cancer cells via epithelial-to-mesenchymal transition through the Wnt/β-catenin pathway[J]. Oncol Rep, 2020, 44( 2): 711- 721. DOI: 10.3892/or.2020.7641. [42] ABECASSIS A, HERMANO E, YIFRACH A, et al. Heparanase contributes to pancreatic carcinoma progression through insulin-dependent glucose uptake[J]. Front Cell Dev Biol, 2023, 11: 1287084. DOI: 10.3389/fcell.2023.1287084. [43] ZHANG H, XU CX, SHI C, et al. Hypermethylation of heparanase 2 promotes colorectal cancer proliferation and is associated with poor prognosis[J]. J Transl Med, 2021, 19( 1): 98. DOI: 10.1186/s12967-021-02770-0. [44] KAYAL Y, BARASH U, NARODITSKY I, et al. Heparanase 2(Hpa2)- a new player essential for pancreatic acinar cell differentiation[J]. Cell Death Dis, 2023, 14( 7): 465. DOI: 10.1038/s41419-023-05990-y. [45] VLODAVSKY I, HILWI M, KAYAL Y, et al. Impact of heparanase-2(Hpa2) on cancer and inflammation: Advances and paradigms[J]. FASEB J, 2024, 38( 10): e23670. DOI: 10.1096/fj.202400286R. [46] CASSINELLI G, TORRI G, NAGGI A. Non-anticoagulant heparins as heparanase inhibitors[J]. Adv Exp Med Biol, 2020, 1221: 493- 522. DOI: 10.1007/978-3-030-34521-1_20. [47] ZHANG YZ, XIONG MJ, CHEN ZX, et al. Design principle of heparanase inhibitors: A combined in vitro and in silico study[J]. ACS Med Chem Lett, 2024, 15( 7): 1032- 1040. DOI: 10.1021/acsmedchemlett.3c00268. [48] ZHANG YZ, CUI LN. Discovery and development of small-molecule heparanase inhibitors[J]. Bioorg Med Chem, 2023, 90: 117335. DOI: 10.1016/j.bmc.2023.117335. [49] RUS A, BOLANOS-GARCIA VM, BASTIDA A, et al. Identification of novel potential heparanase inhibitors using virtual screening[J]. Catalysts, 2022, 12( 5): 503. DOI: 10.3390/catal12050503. [50] LEBSIR N, ZOULIM F, GRIGOROV B. Heparanase-1: From cancer biology to a future antiviral target[J]. Viruses, 2023, 15( 1): 237. DOI: 10.3390/v15010237. [51] de BOER C, ARMSTRONG Z, LIT VAJ, et al. Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo[J]. Proc Natl Acad Sci USA, 2022, 119( 31): e2203167119. DOI: 10.1073/pnas.2203167119. -


 PDF下载 ( 1157 KB)
PDF下载 ( 1157 KB)

 下载:
下载:


