乙型肝炎病毒感染的自然控制和清除
DOI: 10.12449/JCH260102
-
摘要: 乙型肝炎病毒(HBV)是一种独特的嗜肝DNA病毒,在肝细胞核内形成共价闭合环状DNA并可部分整合入宿主基因组,构成病毒持续感染的分子基础。HBV感染复制依赖多种肝细胞富集因子及微环境调控。宿主通过多种方式实现对HBV的自然控制和清除,包括以细胞毒性T细胞及自然杀伤细胞等细胞免疫介导的溶细胞性清除,以干扰素和各类细胞因子驱动的天然免疫与非溶细胞性清除,以及体液免疫中抗体介导的保护与清除。此外,细胞内限制性因子和通路、肝细胞分裂更新及基底硬度等肝脏微环境变化亦共同影响病毒控制和清除的效率与结局。本文深入阐明并动态解析相关机制,将有助于加深理解乙型肝炎慢性化及自愈和治愈的过程,为优化诊疗及研发新型干预策略提供理论基础。Abstract: Hepatitis B virus (HBV) is a unique hepatotropic DNA virus that forms covalently closed circular DNA within the nucleus of hepatocytes and can partially integrate into the host genome, establishing the molecular basis for persistent viral infection. HBV infection and replication depends on multiple hepatocyte-enriched host factors and is modulated by the hepatic microenvironment. The host achieves natural control and clearance of HBV through various mechanisms, including cytolytic elimination mediated by cellular immunity such as cytotoxic T lymphocytes and natural killer cells, innate immunity and noncytolytic clearance driven by interferons and various cytokines, and antibody-mediated protection and clearance as part of humoral immune response. In addition, intracellular restriction factors and pathways, hepatocyte turnover through division and replacement, and changes in the hepatic microenvironment (such as the increase in matrix stiffness) collectively influence the efficiency and outcome of viral control and clearance. This article clarifies and elaborates on related mechanisms, so as to deepen the understanding of HBV chronicity, spontaneous resolution, and cure and provide a theoretical basis for optimizing clinical management and developing novel therapeutic strategies.
-
Key words:
- Hepatitis B Virus /
- Covalently Closed Circular DNA /
- Antiviral Immunity
-
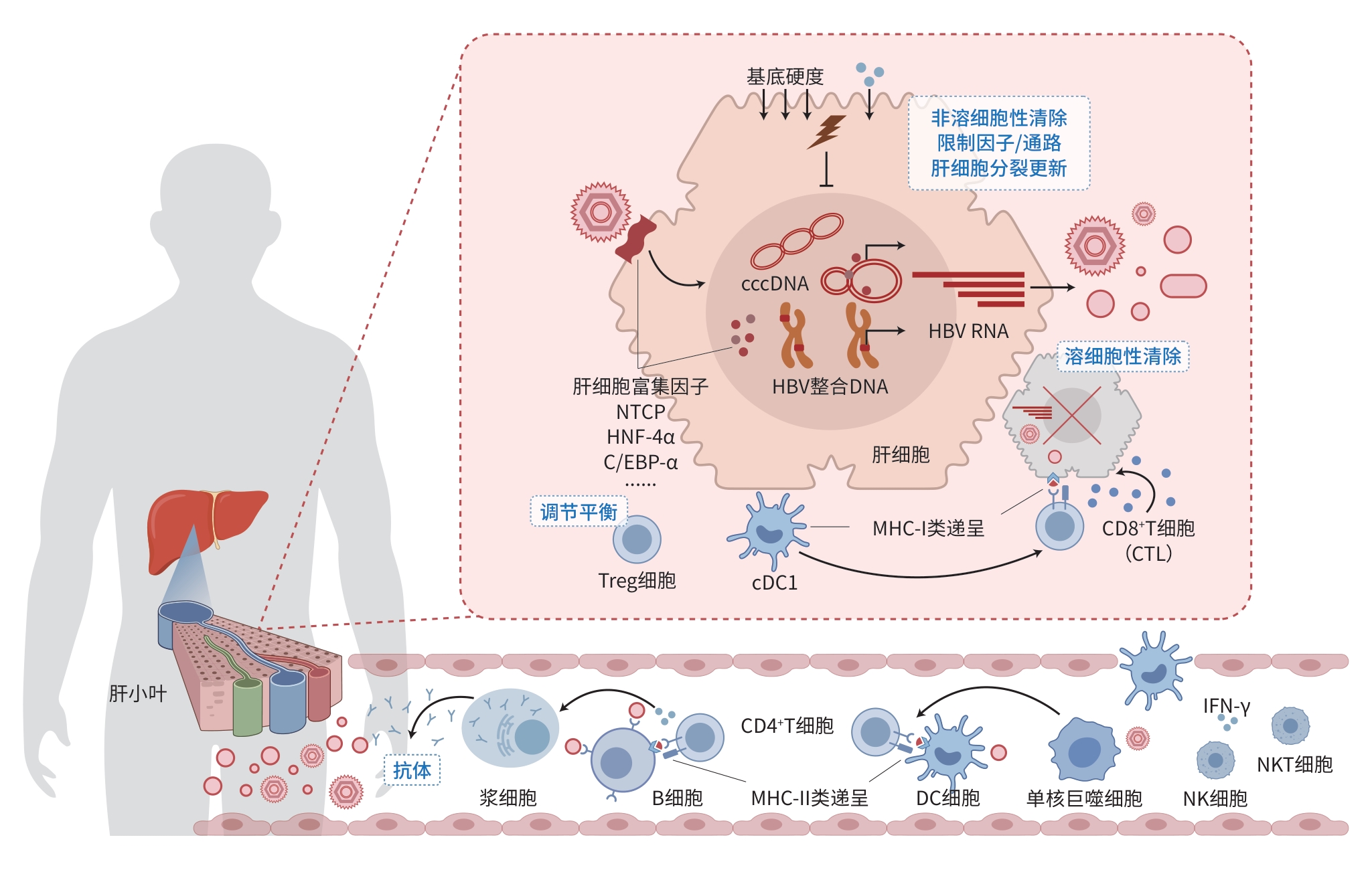
注: HBV是一种嗜肝DNA病毒,感染肝细胞后形成cccDNA和整合DNA,转录产生多种HBV RNA以合成病毒抗原和子代病毒,过程中多种肝细胞富集因子对感染复制的建立与维持具有重要作用。宿主通过溶细胞性、非溶细胞性、限制性因子/通路、肝细胞分裂更新及抗病毒抗体等多种机制控制和清除HBV,期间各类免疫细胞、细胞因子及细胞外基底硬度变化等协同作用。cccDNA,共价闭合环状DNA;HBV,乙型肝炎病毒;NTCP,钠离子-牛磺胆酸共转运蛋白;HNF-4α,肝细胞核因子4α;C/EBP-α,CCAAT增强子结合蛋白α;Treg,调节性T细胞;MHC,主要组织相容性复合体;CTL,细胞毒性T细胞;cDC1,Ⅰ型经典树突状细胞;DC,树突状细胞;IFN-γ,干扰素γ;NKT,自然杀伤T细胞;NK,自然杀伤细胞。
图 1 HBV感染与控制清除机制示意图
Figure 1. Schematic of HBV infection and the mechanisms of viral control and clearance
-
[1] GBD 2019 Hepatitis B Collaborators. Global, regional, and national burden of hepatitis B, 1990-2019: A systematic analysis for the Global Burden of Disease Study 2019[J]. Lancet Gastroenterol Hepatol, 2022, 7( 9): 796- 829. DOI: 10.1016/S2468-1253(22)00124-8. [2] YAN H, ZHONG GC, XU GW, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus[J]. eLife, 2012, 1: e00049. DOI: 10.7554/eLife.00049. [3] WEI L, PLOSS A. Core components of DNA lagging strand synthesis machinery are essential for hepatitis B virus cccDNA formation[J]. Nat Microbiol, 2020, 5( 5): 715- 726. DOI: 10.1038/s41564-020-0678-0. [4] ZOULIM F, CHEN PJ, DANDRI M, et al. Hepatitis B virus DNA integration: Implications for diagnostics, therapy, and outcome[J]. J Hepatol, 2024, 81( 6): 1087- 1099. DOI: 10.1016/j.jhep.2024.06.037. [5] QU BQ, BROWN RJP. Strategies to inhibit hepatitis B virus at the transcript level[J]. Viruses, 2021, 13( 7): 1327. DOI: 10.3390/v13071327. [6] YUAN HF, ZHAO LN, YANG G, et al. HBx-induced HSPB1 is a potential therapeutic target owing to its modulation of HBV cccDNA and hepatic immune responses[J]. J Hepatol, 2025. DOI: 10.1016/j.jhep.2025.09.033.[ Epub ahead of print] [7] YE JY, LI FH, HUA T, et al. Liver mechanosignaling as a natural anti-hepatitis B virus mechanism[J]. Nat Commun, 2024, 15( 1): 8375. DOI: 10.1038/s41467-024-52718-3. [8] WIELAND SF, CHISARI FV. Stealth and cunning: Hepatitis B and hepatitis C viruses[J]. J Virol, 2005, 79( 15): 9369- 9380. DOI: 10.1128/JVI.79.15.9369-9380.2005. [9] MAO RC, ZHANG JM, JIANG D, et al. Indoleamine 2, 3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells[J]. J Virol, 2011, 85( 2): 1048- 1057. DOI: 10.1128/JVI.01998-10. [10] SINHA P, THIO CL, BALAGOPAL A. Intracellular host restriction of hepatitis B virus replication[J]. Viruses, 2024, 16( 5): 764. DOI: 10.3390/v16050764. [11] ZHANG W, CHEN JL, WU M, et al. PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation[J]. Hepatology, 2017, 66( 2): 398- 415. DOI: 10.1002/hep.29133. [12] REN JH, HU JL, CHENG ST, et al. SIRT3 restricts hepatitis B virus transcription and replication through epigenetic regulation of covalently closed circular DNA involving suppressor of variegation 3-9 homolog 1 and SET domain containing 1A histone methyltransferases[J]. Hepatology, 2018, 68( 4): 1260- 1276. DOI: 10.1002/hep.29912. [13] DECORSIÈRE A, MUELLER H, van BREUGEL PC, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor[J]. Nature, 2016, 531( 7594): 386- 389. DOI: 10.1038/nature17170. [14] YAO QY, PENG B, LI C, et al. SLF2 interacts with the SMC5/6 complex to direct hepatitis B virus episomal DNA to promyelocytic leukemia bodies for transcriptional repression[J]. J Virol, 2023, 97( 7): e00328-23. DOI: 10.1128/jvi.00328-23. [15] WANG YX, NIKLASCH M, LIU TT, et al. Interferon-inducible MX2 is a host restriction factor of hepatitis B virus replication[J]. J Hepatol, 2020, 72( 5): 865- 876. DOI: 10.1016/j.jhep.2019.12.009. [16] SHEN ZL, ZHANG SY, GAO ZX, et al. Intrahepatic homeobox protein MSX-1 is a novel host restriction factor of hepatitis B virus[J]. J Virol, 2024, 98( 2): e01345-23. DOI: 10.1128/jvi.01345-23. [17] CHENG ST, HU JL, REN JH, et al. Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx[J]. J Hepatol, 2021, 74( 3): 522- 534. DOI: 10.1016/j.jhep.2020.09.019. [18] ALLWEISS L, GIERSCH K, PIROSU A, et al. Therapeutic shutdown of HBV transcripts promotes reappearance of the SMC5/6 complex and silencing of the viral genome in vivo[J]. Gut, 2022, 71( 2): 372- 381. DOI: 10.1136/gutjnl-2020-322571. [19] KAJINO K, JILBERT AR, SAPUTELLI J, et al. Woodchuck hepatitis virus infections: Very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte[J]. J Virol, 1994, 68( 9): 5792- 5803. DOI: 10.1128/JVI.68.9.5792-5803.1994. [20] GUIDOTTI LG, ANDO K, HOBBS MV, et al. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice[J]. Proc Natl Acad Sci USA, 1994, 91( 9): 3764- 3768. DOI: 10.1073/pnas.91.9.3764. [21] XIA YC, STADLER D, LUCIFORA J, et al. Interferon-γ and tumor necrosis factor-α produced by T cells reduce the HBV persistence form, cccDNA, without cytolysis[J]. Gastroenterology, 2016, 150( 1): 194- 205. DOI: 10.1053/j.gastro.2015.09.026. [22] SHEN F, LI YM, WANG Y, et al. Hepatitis B virus sensitivity to interferon-α in hepatocytes is more associated with cellular interferon response than with viral genotype[J]. Hepatology, 2018, 67( 4): 1237- 1252. DOI: 10.1002/hep.29609. [23] DING JH, YI ZG, ZAI WJ, et al. Illuminating the live-cell dynamics of hepatitis B virus covalently closed circular DNA using the CRISPR-tag system[J]. mBio, 2023, 14( 2): e03550-22. DOI: 10.1128/mbio.03550-22. [24] GEHRING AJ, PROTZER U. Targeting innate and adaptive immune responses to cure chronic HBV infection[J]. Gastroenterology, 2019, 156( 2): 325- 337. DOI: 10.1053/j.gastro.2018.10.032. [25] LI XF, SUN WX, XU XL, et al. Hepatitis B virus surface antigen drives T cell immunity through non-canonical antigen presentation in mice[J]. Nat Commun, 2025, 16( 1): 4591. DOI: 10.1038/s41467-025-59985-8. [26] ANDREATA F, LAURA C, RAVÀ M, et al. Therapeutic potential of co-signaling receptor modulation in hepatitis B[J]. Cell, 2024, 187( 15): 4078- 4094. e 21. DOI: 10.1016/j.cell.2024.05.038. [27] BOSCH M, KALLIN N, DONAKONDA S, et al. A liver immune rheostat regulates CD8 T cell immunity in chronic HBV infection[J]. Nature, 2024, 631( 8022): 867- 875. DOI: 10.1038/s41586-024-07630-7. [28] HEIM K, SAGAR, SOGUKPINAR Ö, et al. Attenuated effector T cells are linked to control of chronic HBV infection[J]. Nat Immunol, 2024, 25( 9): 1650- 1662. DOI: 10.1038/s41590-024-01928-4. [29] LI YY, TANG LB, GUO L, et al. CXCL13-mediated recruitment of intrahepatic CXCR5(+)CD8+ T cells favors viral control in chronic HBV infection[J]. J Hepatol, 2020, 72( 3): 420- 430. DOI: 10.1016/j.jhep.2019.09.031. [30] WANG Q, MICHAILIDIS E, YU YP, et al. A combination of human broadly neutralizing antibodies against hepatitis B virus HBsAg with distinct epitopes suppresses escape mutations[J]. Cell Host Microbe, 2020, 28( 2): 335- 349. e 6. DOI: 10.1016/j.chom.2020.05.010. [31] LE BERT N, SALIMZADEH L, GILL US, et al. Comparative characterization of B cells specific for HBV nucleocapsid and envelope proteins in patients with chronic hepatitis B[J]. J Hepatol, 2020, 72( 1): 34- 44. DOI: 10.1016/j.jhep.2019.07.015. [32] LI J, MA X, XUAN QK, et al. Modulation of monocyte activity by hepatocellular microRNA delivery through HBsAg particles: Implications for pathobiology of chronic hepatitis B[J]. Hepatology, 2025, 81( 3): 990- 1005. DOI: 10.1097/HEP.0000000000000972. [33] PUBLICOVER J, GAGGAR A, JESPERSEN JM, et al. An OX40/OX40L interaction directs successful immunity to hepatitis B virus[J]. Sci Transl Med, 2018, 10( 433): eaah5766. DOI: 10.1126/scitranslmed.aah5766. [34] ZHANG C, LI JS, CHENG YQ, et al. Single-cell RNA sequencing reveals intrahepatic and peripheral immune characteristics related to disease phases in HBV-infected patients[J]. Gut, 2023, 72( 1): 153- 167. DOI: 10.1136/gutjnl-2021-325915. [35] QI RY, FU R, LEI X, et al. Therapeutic vaccine-induced plasma cell differentiation is defective in the presence of persistently high HBsAg levels[J]. J Hepatol, 2024, 80( 5): 714- 729. DOI: 10.1016/j.jhep.2023.12.032. [36] PROTZER U, MAINI MK, KNOLLE PA. Living in the liver: Hepatic infections[J]. Nat Rev Immunol, 2012, 12( 3): 201- 213. DOI: 10.1038/nri3169. [37] XU L, YIN WW, SUN R, et al. Kupffer cell-derived IL-10 plays a key role in maintaining humoral immune tolerance in hepatitis B virus-persistent mice[J]. Hepatology, 2014, 59( 2): 443- 452. DOI: 10.1002/hep.26668. [38] FANG Z, ZHANG Y, ZHU ZQ, et al. Monocytic MDSCs homing to thymus contribute to age-related CD8+ T cell tolerance of HBV[J]. J Exp Med, 2022, 219( 4): e20211838. DOI: 10.1084/jem.20211838. [39] THIMME R, HOFMANN M, BERTOLETTI A, et al. Decoding HBV-specific adaptive immunity: From natural clearance to cure[J]. Gut, 2025. DOI: 10.1136/gutjnl-2025-337129.[ Epub ahead of print] [40] LIM SG, BAUMERT TF, BONI C, et al. The scientific basis of combination therapy for chronic hepatitis B functional cure[J]. Nat Rev Gastroenterol Hepatol, 2023, 20( 4): 238- 253. DOI: 10.1038/s41575-022-00724-5. [41] JI Y, LE BERT N, WONG GL, et al. The impact of hepatitis B surface antigen reduction via small interfering RNA treatment on natural and vaccine(BRII-179)-induced hepatitis B virus-specific humoral and cellular immune responses[J]. Gastroenterology, 2025, 169( 1): 136- 149. DOI: 10.1053/j.gastro.2025.02.016. [42] IANNACONE M, BECCARIA CG, ALLWEISS L, et al. Targeting HBV with RNA interference: Paths to cure[J]. Sci Transl Med, 2025, 17( 805): eadv3678. DOI: 10.1126/scitranslmed.adv3678. [43] HU KY, ZAI WJ, XU MZ, et al. Augmented epigenetic repression of hepatitis B virus covalently closed circular DNA by interferon-α and small-interfering RNA synergy[J]. mBio, 2024, 15( 12): e02415-24. DOI: 10.1128/mbio.02415-24. [44] LIN CR, HUANG Y, RAN N, et al. Therapeutic inhibition of HBsAg and HBV cccDNA through a novel phased combination treatment: Glycine and interferon-α[J]. Gut, 2025, 74( 12): 2035- 2049. DOI: 10.1136/gutjnl-2025-334813. [45] ASHUO A, LIU J, YUAN ZH, et al. Interferon-α for immune modulation in chronic hepatitis B toward functional cure[J]. Viruses, 2025, 17( 10): 1358. DOI: 10.3390/v17101358. -


 PDF下载 ( 898 KB)
PDF下载 ( 898 KB)

 下载:
下载:


